Quantemol Database: Your Trusted Plasma Chemistry Data
Quantemol Database (QDB) is a reliable resource for plasma chemistry information, designed for experts working with complex chemistries. It supports and compares multiple data sets to provide validated chemistry reaction sets compatible with various plasma modelling software packages. Request a Free Trial
Quantemol Database: Your Trusted Plasma Chemistry Data
Quantemol Database (QDB) is a reliable resource for plasma chemistry information, designed for experts working with complex chemistries. It supports and compares multiple data sets to provide validated chemistry reaction sets compatible with various plasma modelling software packages. Request a Free Trial
Quantemol Database: Your Trusted Plasma Chemistry Data
Quantemol Database (QDB) is a reliable resource for plasma chemistry information, designed for experts working with complex chemistries. It supports and compares multiple data sets to provide validated chemistry reaction sets compatible with various plasma modelling software packages. Request a Free Trial

Use Your Data with Confidence
QDB uses trustworthy, validated data and chemistry sets that are monitored regularly, so you can be confident in your results.

All of Your Data in One Place
QDB supports streamlined comparisons of multiple data sets, such as: electron scattering, heavy particle collisions and surface chemistry.
Save Precious Time
QDB’s easy-to-use tools will help you to assemble the chemistry set you need in minutes. It intuitively exports data in flexible formats & avoids human error.
Connect with Our Team
We offer ongoing data updates and support, so you don’t have to struggle alone!

Use Your Data with Confidence
QDB uses trustworthy, validated data and chemistry sets that are monitored regularly, so you can be confident in your results.

All of Your Data in One Place
QDB supports streamlined comparisons of multiple data sets, such as: electron scattering, heavy particle collisions and surface chemistry.
Save Precious Time
QDB’s easy-to-use tools will help you to assemble the chemistry set you need in minutes. It intuitively exports data in flexible formats & avoids human error.
Connect with Our Team
We offer ongoing data updates and support, so you dont have to struggle alone!
Visit our QDB website for more information →
*Most Customers Choose Gold*
↓
Free Standard Membership
- Search the database for reactions and species
- View flexible cross section graphs
- Download up to 20 datasets/month
Gold Membership
- Unlimited dataset downloads
- Search and compare cross sections (same species; cross-species comparison coming soon)
- Access 42+ pre-assembled chemistry sets
- Download formats compatible with COMSOL, STAR CCM+, CHEMKIN, HPEM, VizGlow
- Download:
- Unlimited individual reactions
- 20 full chemistry sets/month
- 20 dynamic sets/month
- Includes monthly renewal
Additional Features:
- Downloadable reference list for cited data
- Plasma expert support to customise chemistry sets
- Help building your own set
- Leave and read user feedback
- Q-VT API access
- Access to:
- Surface chemistry data
- Boltzmann solver (cloud-based + data download)
- Global Plasma Model with:
- Custom surface sticking/return probabilities
- Downloadable outputs (final + time-dependent densities, full reaction list with rate coefficients)
- Pulsed plasma option (dual power levels)
- Save & restore dynamic sets online
Quantemol Global Model Software
Platinum Membership
(included with your 1-year membership)
- Request cross section data for up to 6 molecules/ions anytime during your membership—no need for repeated funding approvals
- Includes a comprehensive report and delivery in your required format
Cross Sections Covered:
- Elastic
- Electronic excitation
- Super-elastic
- Quenching
- Electron impact dissociation
- Dissociative electron attachment (estimated)
- Momentum transfer
- Ionisation (all energies)
- Rotational excitation
- Vibrational (for diatomic molecules: N₂, FO, CF, etc.)
- Branching ratios for ionisation/dissociation (estimated)
Molecule Limits:
- Up to 20 atoms (preferably lighter than Ar): all above processes
- Over 20 atoms: selected processes only
- Dissociative attachment
- Elastic
- Electronic excitation (to first excited state)
- Ionisation
All Gold Membership benefits are included
Visit our QDB website for more information →
*Most Customers Choose Gold*
↓
Free Standard Membership
- Search the database for reactions and species
- View flexible cross section graphs
- Download up to 20 datasets/month
Gold Membership
- Unlimited dataset downloads
- Search and compare cross sections (same species; cross-species comparison coming soon)
- Access 42+ pre-assembled chemistry sets
- Download formats compatible with COMSOL, STAR CCM+, CHEMKIN, HPEM, VizGlow
- Download:
- Unlimited individual reactions
- 20 full chemistry sets/month
- 20 dynamic sets/month
- Includes monthly renewal
Additional Features:
- Downloadable reference list for cited data
- Plasma expert support to customise chemistry sets
- Help building your own set
- Leave and read user feedback
- Q-VT API access
- Access to:
- Surface chemistry data
- Boltzmann solver (cloud-based + data download)
- Global Plasma Model with:
- Custom surface sticking/return probabilities
- Downloadable outputs (final + time-dependent densities, full reaction list with rate coefficients)
- Pulsed plasma option (dual power levels)
- Save & restore dynamic sets online
Quantemol Global Model Software
Platinum Membership
(included with your 1-year membership)
- Request cross section data for up to 6 molecules/ions anytime during your membership—no need for repeated funding approvals
- Includes a comprehensive report and delivery in your required format
Cross Sections Covered:
- Elastic
- Electronic excitation
- Super-elastic
- Quenching
- Electron impact dissociation
- Dissociative electron attachment (estimated)
- Momentum transfer
- Ionisation (all energies)
- Rotational excitation
- Vibrational (for diatomic molecules: N₂, FO, CF, etc.)
- Branching ratios for ionisation/dissociation (estimated)
Molecule Limits:
- Up to 20 atoms (preferably lighter than Ar): all above processes
- Over 20 atoms: selected processes only
- Dissociative attachment
- Elastic
- Electronic excitation (to first excited state)
- Ionisation
All Gold Membership benefits are included
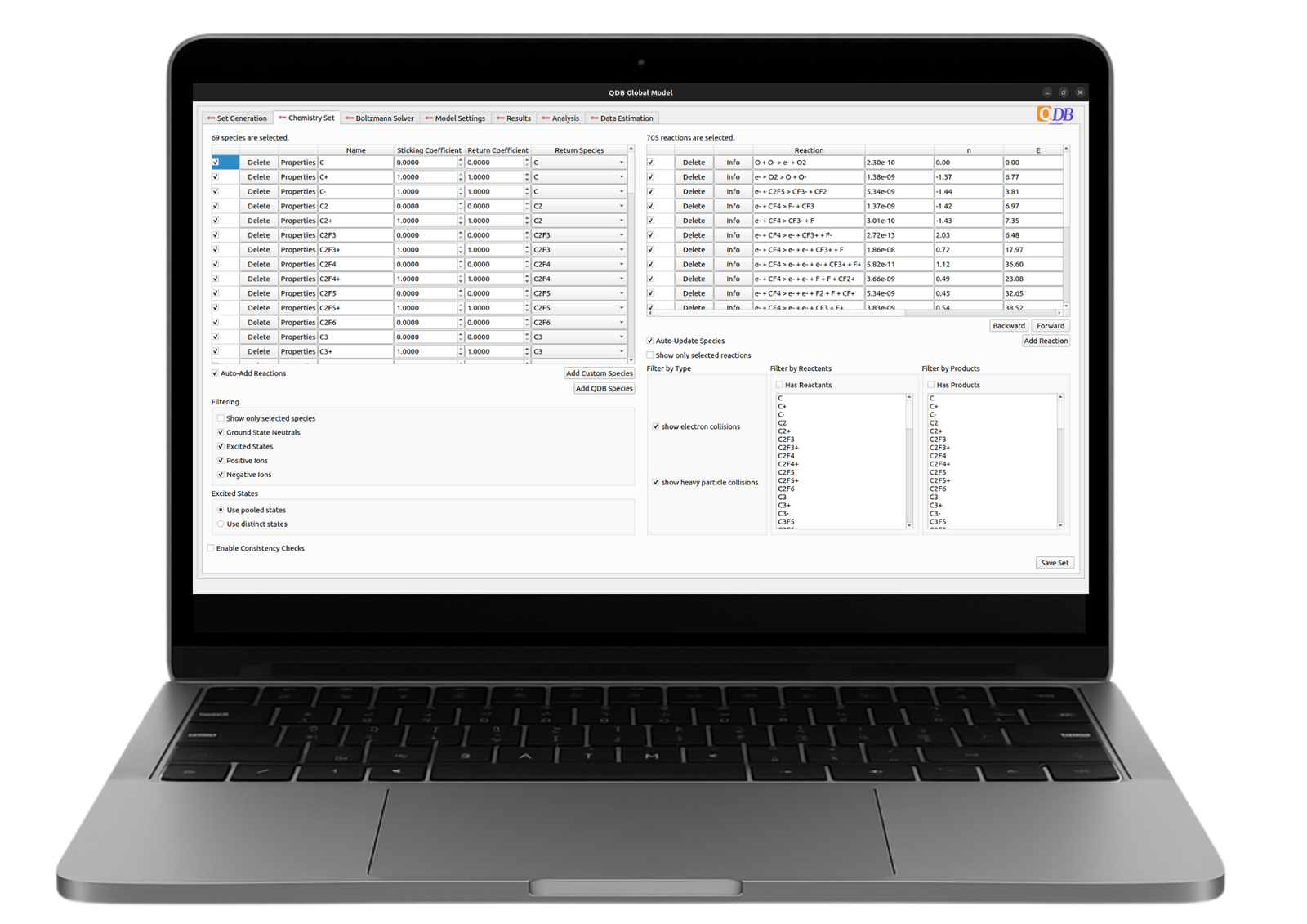
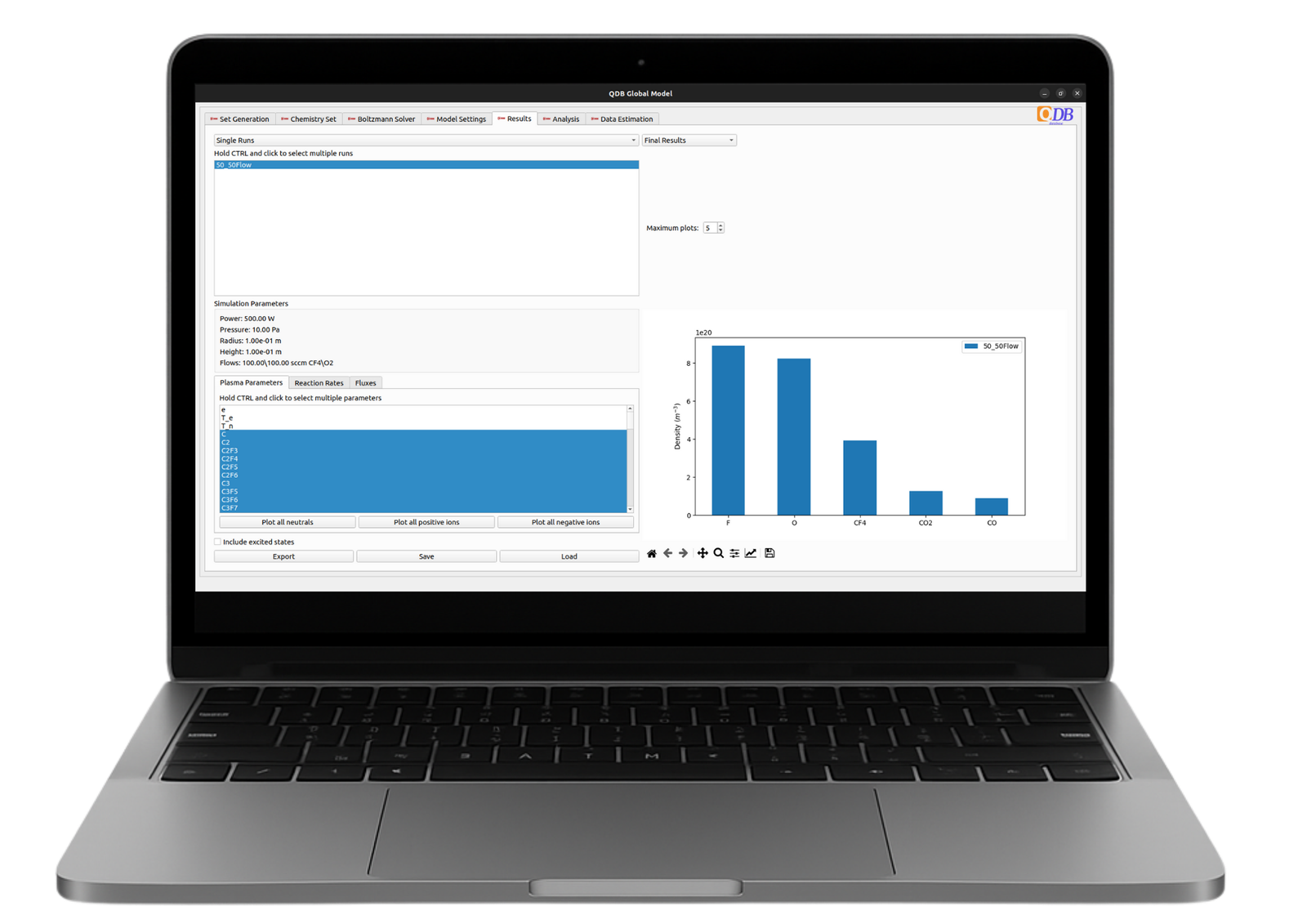
Included with your Quantemol-DB package, the Quantemol Global Model is a powerful tool for calculating reactor-averaged particle densities and electron temperature based on your plasma process parameters.
You can test the online version is free version on pre assembled chemistry sets. A more advanced desktop app is exclusive to QDB Gold and Platinum Members.
- User-friendly and intuitive interface.
- Flexible chemistry selection: linked via API to QDB (Gold/Platinum Members).
- Generate new or download existing chemistry sets
- Freely populate with your preferred species.
- Self-consistency checks for chemistry sets.
- Options for pulsed discharge and parameter variation studies.
- Convenient plotting and results comparison.
- Stand-alone Boltzmann Solver and integration with the Global Model.
- Analysis tools: extract/plot rates, chemistry set reduction/ optimisation.
- Integration with machine learning models
- Import/export input files for HPEM, CHEMKIN, COMSOL, etc.
- Stand-alone Surface Model and integration with Global Model.
- Analysis: visualising reaction pathways.
Included with your Quantemol-DB package, the Quantemol Global Model is a powerful tool for calculating reactor-averaged particle densities and electron temperature based on your plasma process parameters.
You can test the online version is free version on pre assembled chemistry sets. A more advanced desktop app is exclusive to QDB Gold and Platinum Members.
- User-friendly and intuitive interface.
- Flexible chemistry selection: linked via API to QDB (Gold/Platinum Members).
- Generate new or download existing chemistry sets
- Freely populate with your preferred species.
- Self-consistency checks for chemistry sets.
- Options for pulsed discharge and parameter variation studies.
- Convenient plotting and results comparison.
- Stand-alone Boltzmann Solver and integration with the Global Model.
- Analysis tools: extract/plot rates, chemistry set reduction/ optimisation.
- Integration with machine learning models
- Import/export input files for HPEM, CHEMKIN, COMSOL, etc.
- Stand-alone Surface Model and integration with Global Model.
- Analysis: visualising reaction pathways.


Quantemol has developed a machine learning model to quickly estimate reaction rate coefficients for heavy particle collisions. Trained on ~10,000 data points from trusted sources (QDB, NFRI, KIDA, UDfA), the model uses common species properties like molar mass, charge, enthalpy, dipole moment, and composition.
The model is powered by a voting regressor, combining:
- Support Vector Regression
- Random Forest
- Gradient Boosted Trees
- k-Nearest Neighbours
Performance:
In testing on over 1,000 reactions, 87% of predictions were within one order of magnitude of reported values. Please note, some predictions may have larger errors.
Note: Machine learning offers fast estimates but may not match the accuracy of quantum chemistry methods. Use with informed judgment.
Use this tool to fill data gaps in your chemistry sets and accelerate model development.
Features:
➱Estimator for Rate Coefficients:
Quickly fill in missing data for your plasma simulations. This tool rapidly estimates reaction rate coefficients for heavy particle collisions, helping you build more complete chemistry sets for your research. This feature is available to all members.
➱Estimator for Mass Spectra & Partial Cross Sections:
Gain deeper insights into electron impact ionisation processes. This method predicts mass spectra to help estimate previously unavailable partial ionisation cross sections, enriching your datasets for more accurate plasma modelling.
➱Diffusion Coefficient Estimators:
Obtain crucial transport data more efficiently with machine learning-based estimations for binary diffusion coefficients. Allowing faster parameterisation of your plasma models, particularly when experimental data for diffusion coefficients are scarce or difficult to obtain.
Explore more about machine learning on the QDB Website

Please note:
- Elements are case-sensitive, e.g. Ar,O,CH4,HCl
- Ions are specified by + or -, eventually followed by the charge number, e.g. H+, Cl-, Ar+2
- Species are separeted by ‘ + ‘ and reactants and products are separated by ‘ -> ‘. Please mind the spaces between the species and these symbols.
- Reaction rate coefficient only works for two reactant-product pairs e.g ‘Ar+ + H2O -> Ar + H2O+’ or for reactions with three products e.g ‘O3 + HO2 -> O2 + O2 + OH’
Machine Learning References
Alves, L. et al. (2022) ‘The 2021 release of the Quantemol database (QDB) of plasma chemistries and reactions’, Plasma Sources Science and Technology, 31(9), p. 095020.
Biau, G. and Devroye, L. (2015) Lectures on the Nearest Neighbor Method. New York: Springer.
Boser, B. E., Guyon, I. M. and Vapnik, V. N. (1992) ‘A training algorithm for optimal margin classifiers’, in Proceedings of the fifth annual workshop on Computational learning theory – COLT ’92 (Pittsburgh, Pennsylvania, United States), ACM Press, pp. 144–152.
Breiman, L. (1997) Arcing the edge. Available at: https://statistics.berkeley.edu/sites/default/files/tech-reports/486.pdf
Breiman, L. (ed.) (1998) Classification and regression trees. 1st edn. Boca Raton, Fla.: Chapman & Hall/CRC.
McElroy, D., Walsh, C., Markwick, A. J., Cordiner, M. A., Smith, K. and Millar, T. J. (2013) ‘The UMIST database for astrochemistry 2012’, Astronomy & Astrophysics, 550, p. A36.
Park, J. H., Choi, H., Chang, W. S., Chung, S. Y., Kwon, D. C., Song, M. Y. and Yoon, J. S. (2020) ‘A new version of the plasma database for plasma physics in the data center for plasma properties’, Applied Science and Convergence Technology, 29(4), pp. 69–74.
Wakelam, V. et al.. (2012) ‘A kinetic database for Astrochemistry (KIDA)’, The Astrophysical Journal Supplement Series, 199(1), p. 21.
Please use this reference to cite the data obtained with the model: K. M. Lemishko, M. Hanicinec, S. Mohr, A. Nelson and J. Tennyson, “Machine learning for plasma reaction kinetic data estimation,” 2024 IEEE International Conference on Plasma Science (ICOPS), Beijing, China, 2024.
Quantemol has developed a machine learning model to quickly estimate reaction rate coefficients for heavy particle collisions. Trained on ~10,000 data points from trusted sources (QDB, NFRI, KIDA, UDfA), the model uses common species properties like molar mass, charge, enthalpy, dipole moment, and composition.
The model is powered by a voting regressor, combining:
- Support Vector Regression
- Random Forest
- Gradient Boosted Trees
- k-Nearest Neighbours
Performance:
In testing on over 1,000 reactions, 87% of predictions were within one order of magnitude of reported values. Please note, some predictions may have larger errors.
Note: Machine learning offers fast estimates but may not match the accuracy of quantum chemistry methods. Use with informed judgment.
Use this tool to fill data gaps in your chemistry sets and accelerate model development.
Features:
➱Estimator for Rate Coefficients:
Quickly fill in missing data for your plasma simulations. This tool rapidly estimates reaction rate coefficients for heavy particle collisions, helping you build more complete chemistry sets for your research. This feature is available to all members.
➱Estimator for Mass Spectra & Partial Cross Sections:
Gain deeper insights into electron impact ionisation processes. This method predicts mass spectra to help estimate previously unavailable partial ionisation cross sections, enriching your datasets for more accurate plasma modelling.
➱Diffusion Coefficient Estimators:
Obtain crucial transport data more efficiently with machine learning-based estimations for binary diffusion coefficients. Allowing faster parameterisation of your plasma models, particularly when experimental data for diffusion coefficients are scarce or difficult to obtain.
Explore more about machine learning on the QDB Website


Please note:
- Elements are case-sensitive, e.g. Ar,O,CH4,HCl
- Ions are specified by + or -, eventually followed by the charge number, e.g. H+, Cl-, Ar+2
- Species are separeted by ‘ + ‘ and reactants and products are separated by ‘ -> ‘. Please mind the spaces between the species and these symbols.
- Reaction rate coefficient only works for two reactant-product pairs e.g ‘Ar+ + H2O -> Ar + H2O+’ or for reactions with three products e.g ‘O3 + HO2 -> O2 + O2 + OH’
Machine Learning References
Alves, L. et al. (2022) ‘The 2021 release of the Quantemol database (QDB) of plasma chemistries and reactions’, Plasma Sources Science and Technology, 31(9), p. 095020.
Biau, G. and Devroye, L. (2015) Lectures on the Nearest Neighbor Method. New York: Springer.
Boser, B. E., Guyon, I. M. and Vapnik, V. N. (1992) ‘A training algorithm for optimal margin classifiers’, in Proceedings of the fifth annual workshop on Computational learning theory – COLT ’92 (Pittsburgh, Pennsylvania, United States), ACM Press, pp. 144–152.
Breiman, L. (1997) Arcing the edge. Available at: https://statistics.berkeley.edu/sites/default/files/tech-reports/486.pdf (Accessed: 22 July 2025).
Breiman, L. (ed.) (1998) Classification and regression trees. 1st edn. Boca Raton, Fla.: Chapman & Hall/CRC.
McElroy, D., Walsh, C., Markwick, A. J., Cordiner, M. A., Smith, K. and Millar, T. J. (2013) ‘The UMIST database for astrochemistry 2012’, Astronomy & Astrophysics, 550, p. A36.
Park, J. H., Choi, H., Chang, W. S., Chung, S. Y., Kwon, D. C., Song, M. Y. and Yoon, J. S. (2020) ‘A new version of the plasma database for plasma physics in the data center for plasma properties’, Applied Science and Convergence Technology, 29(4), pp. 69–74.
Wakelam, V. et al.. (2012) ‘A kinetic database for Astrochemistry (KIDA)’, The Astrophysical Journal Supplement Series, 199(1), p. 21.
Please use this reference to cite the data obtained with the model: K. M. Lemishko, M. Hanicinec, S. Mohr, A. Nelson and J. Tennyson, “Machine learning for plasma reaction kinetic data estimation,” 2024 IEEE International Conference on Plasma Science (ICOPS), Beijing, China, 2024.
“The money we saved pays for the subscription to QDB about 100 times over. We are quite satisfied!”
“We are fully satisfied with your software and support. We are convinced it is absolutely beneficial to our business.”
“The money we saved pays for the subscription to QDB about 100 times over. We are quite satisfied!”
“We are fully satisfied with your software and support. We are convinced it is absolutely beneficial to our business.”


